Abstract
Background. Epithelial cells are the first barrier to any microbial invasion. Finding a safe and affordable substance to stimulate the innate immune response of epithelial cells is one of the main challenges immunologists and vaccine manufacturers are facing.
Objectives. This study aimed to show the comparative effect of sterile bacterial secretion (SBS) and Pseudomonas aeruginosa bacterial cell isolates obtained from burn wound infections on the ability of human epithelial cells (HECs) to produce interleukin (IL)-1β and tumor necrosis factor alpha (TNF-α) in vitro.
Materials and methods. The HEC cultures were exposed to P. aeruginosa 8 (Pa 8), Pa 2 and Pa 1 bacterial cells (isolated from burn wound infections). The other 3 groups of HECs were exposed to 50 μL of sterile, endotoxin-free SBS of Pa 8, Pa 2 and Pa 1. The time course of changes in IL-1β mRNA, TNF-α mRNA, IL-1β, and TNF-α was examined.
Results. Moderate (p < 0.05) elevations of IL-1β mRNA in HECs and IL-1β protein in the supernatant of the HEC culture were observed following exposure to SBS of Pa 8, Pa 2 and Pa 1 at most time points. High elevation (p < 0.05) of IL-1β was seen in the supernatant of the HEC culture that was exposed to bacterial cells (Pa 8, Pa 2 and Pa 1). Similar results were found when TNF-α mRNA was measured in HECs and TNF-α in the supernatant of the HEC cultures after exposure to bacterial cells (Pa 8, Pa 2 and Pa 1) and the SBS of Pa 8, Pa 2 and Pa 1.
Conclusion. This is the first time that the capacity of SBS to generate epithelial cell pro-inflammatory cytokines in vitro has been shown. In other words, SBS enhanced a nonspecific immune response, which opens the door to the possibility of using SBS from P. aeruginosa as an adjuvant in the future.
Keywords: human epithelial cells (HECs), interleukin-1, Pseudomonas aeruginosa, tumor necrosis factor alpha, sterile bacterial secretion (SBS)
Background
Pseudomonas aeruginosa is a highly prevalent pathogen associated with several infectious diseases.1 Infections with these bacterial isolates have a wide range of illness severity. For the most part, persistent infection with this bacterium creates a distinctive phenotype enclosed by a thick alginate of capsular polysaccharide.2 This helps the bacteria survive in the lungs of the host and leads to reduced immune responses against P. aeruginosa, which ultimately results in severe respiratory tract infections.3
Pseudomonas aeruginosa has a genetic adaptation that enables it to persist in the body through its ability to evade the immune system and resist antimicrobial agents.4 In addition, it was found that a number of P. aeruginosa isolates have the ability to secrete substances that enable the bacteria to penetrate the first lines of defense of the respiratory system.5 Previous studies have shown that the secretions of bacterial proteolytic enzymes destroy antimicrobial peptides and proteins, which helps the bacteria overcome the host’s innate immune system.6 Other studies have confirmed the ability of P. aeruginosa to stimulate the innate immune system and pro-inflammatory immune system through its structural proteins and surface proteins (i.e., flagella and pili). Further research has shown that bacterial secretions from P. aeruginosa may help stimulate the pro-inflammatory immune system.7
Epithelial cells are the first barrier in the body in general and in the respiratory system in particular. They protect the body from exposure to chemicals and microorganisms.7 These cells work to prevent the entry of pathogenic microorganisms that cause respiratory system infections. Several previous studies have shown the function of human epithelial cells (HECs) in the mucosal innate response of the immune system through their ability to engulf pathogenic microorganisms, as well as secrete mucus substances that contribute to reducing the adhesion of microorganisms and antibacterial substances.8 Other studies have shown the ability of these cells to secrete different pro-inflammatory cytokines (interleukin (IL)-1β and IL-6, and tumor necrosis factor-alpha (TNF-α)) and chemokines (IL-8) after exposure to bacterial cells, which enables the HECs to have a significant effect on the pro-inflammatory immune response.9
This study aimed to demonstrate the effect of extracellular secretions from P. aeruginosa in growth medium on the ability of epithelial cells to produce pro-inflammatory cytokines in vitro.
Materials and methods
Isolation and identification of bacteria
The standard method developed by Zgair et al. was followed.9 Wound swabs were collected from 110 patients suffering from burn wound infections and treated at the Baghdad Teaching Hospital in Baghdad, Iraq. All individuals gave written informed consent to participate in the study. The patients did not receive any antibiotics for 2 days before sample collection. The swabs were subjected to an asparagine broth enrichment medium to enhance P. aeruginosa growth, and incubated for 48 h at 37°C with vigorous shaking at 200 rpm. A loopful of bacterial suspension was streaked onto asparagine plates containing 1.5% agar (HiMedia, Mumbai, India) and incubated at 37°C until colonies developed.9 A VITEK 2 DensiCHEK fluorescence instrument system with an ID-GNB card (bioMérieux, Marcy-l’Étoile, France) was used to identify the isolates of P. aeruginosa.10
The study was conducted following approval from the Human Ethical Committee of the University of Baghdad, Iraq (Reference No. HS-212, April 1, 2020).
Preparation of bacterial suspension
Pseudomonas aeruginosa clinical isolates (PAC) isolated from infected wounds were grown in Luria−Bertani (LB) broth (HiMedia) at 37°C for 18 h. Bacterial cell pellets were collected by centrifugation (5000 g for 20 min at 4°C). The collected pellets were washed 3 times with phosphate-buffered saline (PBS) (0.01 M, pH 7.2). The final bacterial counts were adjusted to 2 × 107 colony forming units (CFU)/mL with PBS.11
Preparation of sterile bacterial secretion
The PAC and P. aeruginosa environmental isolate (PAE) were grown overnight in LB broth at 37°C (18 h). The supernatants of bacterial growth from the LB broth (sterile bacterial secretion (SBS)) were obtained by centrifugation (5000 g for 20 min at 4°C) and were filtrated using a special Merck Millipore filter (MF-Millipore™ Membrane Filter, 0.22 µm pore size; Merck Millipore, Waltham, USA). The sterility of the supernatants (SBS) was checked by culturing on LB agar plates.
Oral human epithelial cell culture
The HEC samples were obtained from 4 healthy adult donors (2 females and 2 males) aged 30–35 years. The samples were collected from the oral mucosa by scratching the inner surface of the mouth with sterile sticks. The collected mucus was washed with NaCl (0.15 M, pH 7.1) supplemented with 500 U/mL penicillin and 500 μg/mL streptomycin (Life Technologies, Carlsbad, USA) at 100 g (10 min, 4°C). The final volume was adjusted to 2 mL with NaCl (0.15 M, pH 7.1). The epithelial cell suspension was placed onto different dilutions of Percoll (Sigma-Aldrich, St. Louis, USA) (30%, 40%, 50%, and 60% of stock isotonic Percoll in the final solution). It was determined using a light microscope (model DM300; Leica Microsystems, Wetzlar, Germany) that the concentration of 40% of the dilute yielded the highest percentage of epithelial cells. Zgair’s standard method was followed to prepare the standard cell culture medium of RPMI 1640 (fetal bovine serum (FBS); Sigma-Aldrich) with all supplements.12 This medium was used to re-suspend the collected epithelial cell layer. The suspension was washed 3 times with the standard cell culture medium. The number of epithelial cells was adjusted to 5 × 105 cells/mL using the culture medium. The trypan blue dye exclusion method showed that more than 90% of the cells were viable.
Experiment
The standard number of epithelial cells (5 × 105 viable cells/mL) was re-cultured under standard cell culture conditions.12 The cell cultures were divided into 6 groups of cell culture tubes (NuncTM) (pluriSELECT, Leipzig, Germany). Then, 100 μL of 107 CFU/mL of P. aeruginosa 8 (Pa 8), Pa 6 and Pa 1 were added to culture tubes 1, 2 and 3, respectively, while 100 μL of SBS (Pa 8), SBS (Pa 6) and SBS (Pa 1) were added to culture tubes 4, 5 and 6, respectively. Phosphate-buffered saline was added to the epithelial cell culture as the control subgroup.
Levels of IL-1β and TNF-α
In a next step, 200 μL of the supernatants of the tissue cultures of different groups were collected at different time intervals (0 h, 1 h, 6 h, and 24 h) after stimulation with either bacterial cells or bacterial SBS. The collected supernatants were centrifuged at 600 g for 5 min and filtered (MF-Millipore™ Membrane Filter, 0.2 µm). The prepared supernatants were stored at −20°C until used for enzyme-linked immunosorbent assay (ELISA). The levels of pro-inflammatory cytokines were measured using human TNF-α and human IL-1β ELISA kits (Koma Biotech Inc., Seoul, South Korea) at a wavelength of 450 nm following the manufacturer’s instructions.13
IL-1β and TNF-α mRNA gene expression
Quantitative real-time polymerase chain reaction (RT-PCR) was performed using the ABI Prism 7700 Sequence Detection System (Applied Biosystems, Waltham, USA). RNA was isolated and purified from HECs pelleted from tissue culture at different time intervals (0 h, 1 h, 6 h, 24 h, 48 h, and 72 h) following exposure to either bacterial cells or bacterial SBS using TRIzol reagent (Thermo Fisher Scientific, Waltham, USA). For cDNA synthesis, 1 µg of total RNA was transcribed with TaqMan Reverse Transcription Reagent (Thermo Fisher Scientific) using random hexamers. The PCR primers of IL-1β, TNF-α and G3PDH were designed, and RT-PCR was performed as described previously.14, 15, 16, 17 Quantitative RT-PCR for IL-1β and TNF-α was performed normalized to the copies of GAPDH mRNA from the same sample. The experiments were conducted in duplicate, and the results were calculated as mean values.
Statistical analyses
Origin 8 software (OriginLab Corporation, Northampton, USA) was used to perform the statistical analyses. The data were expressed as mean ± standard error (M ±SE). The differences were evaluated using Student’s t-test and one-way analysis of variance (ANOVA). The p-values <0.05 were considered statistically significant.
Results
Psedomonas aeruginosa isolation
and identification
Briefly, 110 infected wound samples were collected from hospitalized patients. Ten isolates of P. aeruginosa were isolated and identified. Isolates of Pa 8, Pa 6 and Pa 1 were selected to conduct further experiments, which included evaluating the ability of bacterial cells and their SBS to stimulate epithelial cell cultures to produce IL-1β and TNF-α.
Pseudomonas aeruginosa and SBS
stimulate HECs to produce IL-1β in vitro
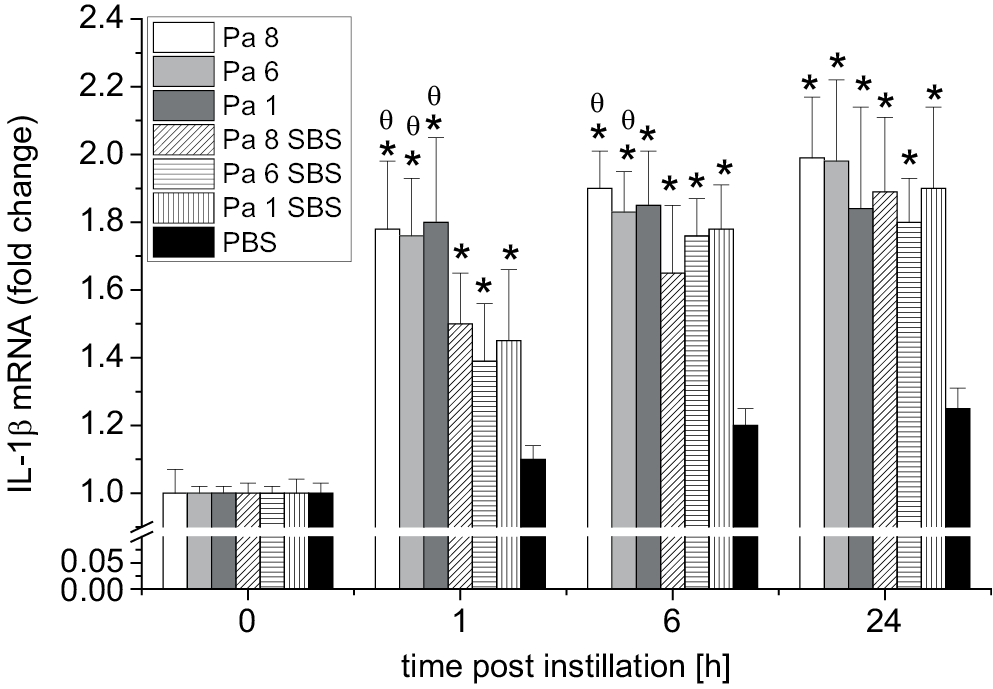
The IL-1β levels in the HEC cultures after exposure to bacterial cells of P. aeruginosa (Pa 8, Pa 6 and Pa 1) and SBS of P. aeruginosa (Pa 8, Pa 6 and Pa 1) (test groups) and the HEC culture exposed to PBS (control group) were measured at different time intervals. Figure 1 shows the fold change in mRNA expression of IL-1β. Significant elevation of IL-1β expression was seen as early as 1 h after exposure to bacterial cells of P. aeruginosa (Pa 8, Pa 6 and Pa 1) and SBS of P. aeruginosa (Pa 8, Pa 6 and Pa 1) compared to the IL-1β expression of the control group (HECs exposed to PBS only). The results indicated that the ability of bacterial cells to stimulate gene expression was higher than the ability of their SBS to stimulate IL-1β expression 1 h and 6 h post-exposure.
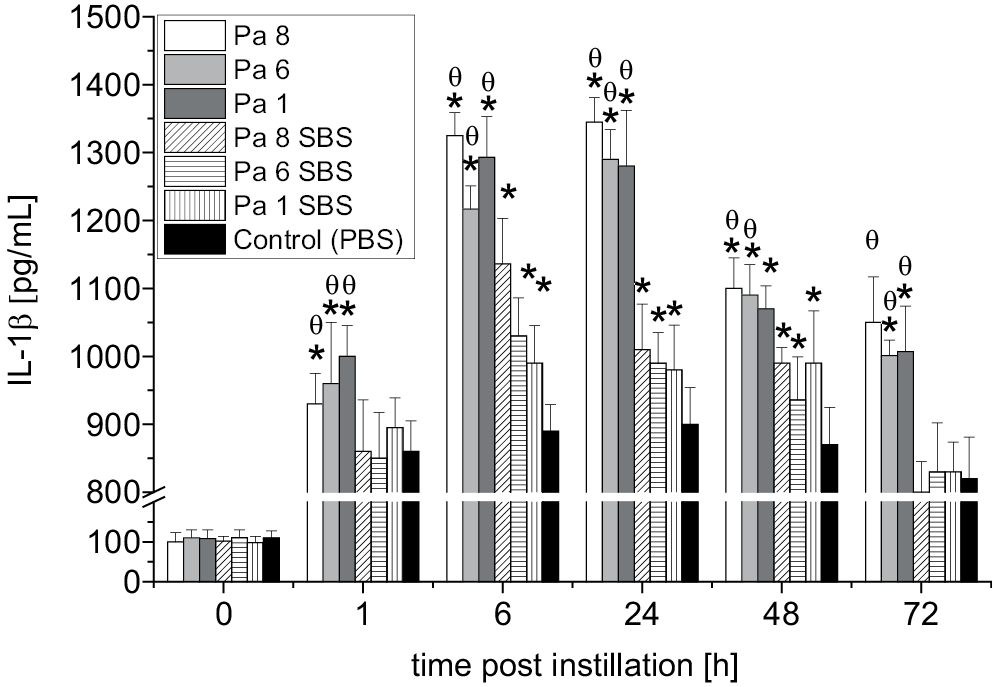
Figure 2 shows the level of IL-1β post-exposure to either bacterial cells or bacterial SBS. A significant elevation of IL-1β was seen as early as 1 h after exposure to bacterial cells of P. aeruginosa (Pa 8, Pa 6 and Pa 1) and SBS of P. aeruginosa (Pa 8, Pa 6 and Pa 1) compared to the IL-1β level of the control group (HECs exposed to PBS only) (p < 0.05). The results indicated that the IL-1β level in the HEC culture that was exposed to bacterial cells was higher than the IL-1β level post-exposure to bacterial SBS (p < 0.01) at all time points. The highest levels of IL-1β were observed 6 h and 24 h after exposure to bacterial cells. The maximum concentrations of IL-1β in the HEC culture post-exposure to SBS were observed 6 h, 24 h and 48 h post-exposure, with only slight differences related to which bacterial strain the SBS was prepared from. The IL-1β levels declined over time; the lowest IL-1β level was observed 72 h post-exposure to bacterial cells and their SBS. At 72 h, the IL-1β levels in the supernatants of the HEC cultures that were exposed to bacterial cells were higher (p < 0.05) than the concentrations of IL-1β in the supernatants of the HEC cultures that were exposed to SBS and PBS (control). At 72 h, there was no significant difference between the concentrations of IL-1β in the HEC cultures that were stimulated with SBS and the IL-1β levels in the HEC culture that was stimulated with PBS (p > 0.05).
Psedomonas aeruginosa and SBS
stimulate HECs to produce TNF-α in vitro
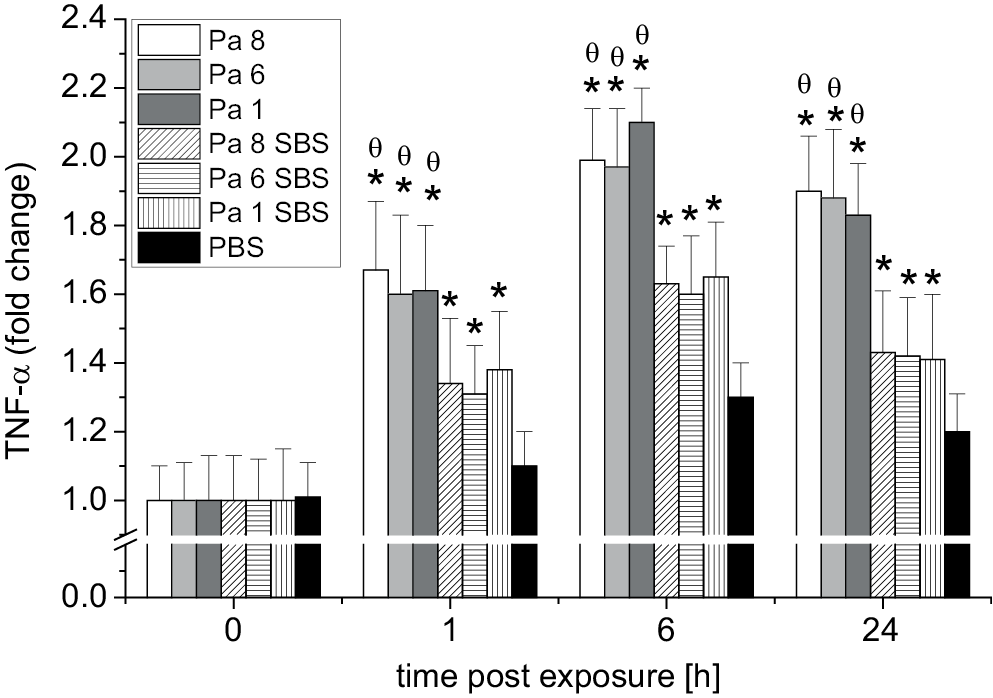
Similarly, the TNF-α levels in the oral HEC cultures following exposure to bacterial cells of P. aeruginosa (Pa 8, Pa 6 and Pa 1) (test group), SBS of P. aeruginosa (Pa 8, Pa 6 and Pa 1) (test group) and PBS (control group) at different time intervals were measured. Figure 1 shows the fold change in TNF-α mRNA expression. A significant rise in TNF-α expression was observed as early as 1 h post-exposure to P. aeruginosa bacterial cells (Pa 8, Pa 6 and Pa 1) and SBS of P. aeruginosa (Pa 8, Pa 6 and Pa 1) compared to the TNF-α expression in the control group (HEC culture exposed to PBS only). The results showed that the ability of bacterial cells to stimulate TNF-α gene expression was higher than the ability of their SBS to stimulate TNF-α gene expression 1 h and 6 h post-exposure (Figure 3).
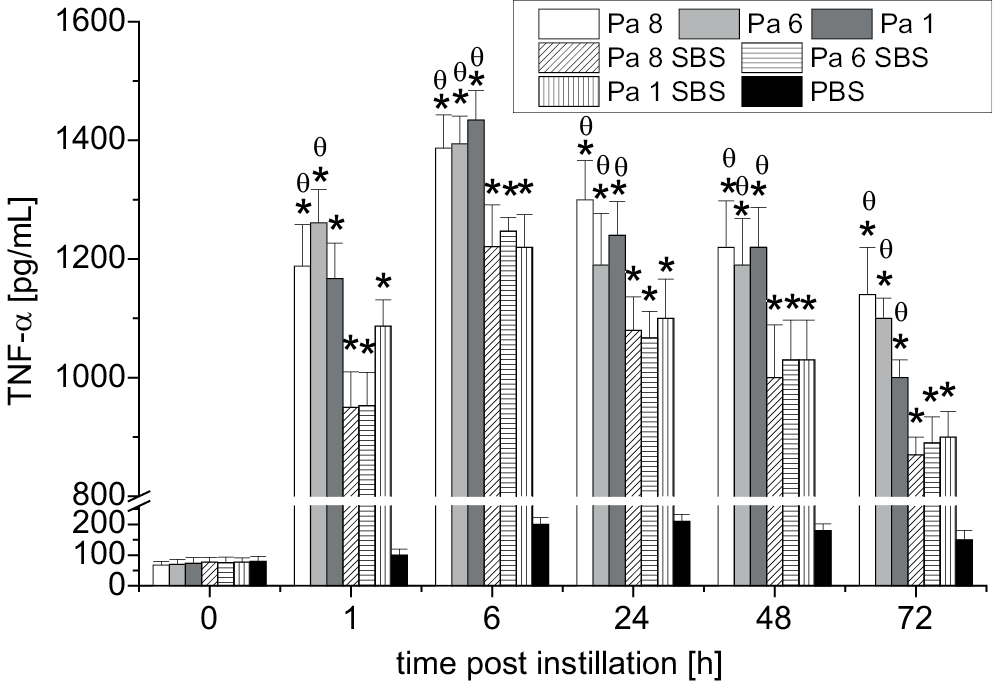
The TNF-α levels in the HEC cultures following exposure to either bacterial cells or bacterial SBS were evaluated. Figure 4 illustrates that significant production of TNF-α was seen as early as 1 h after exposure compared to the TNF-α level in the control group HEC culture (exposed to PBS only). At that time point, significant (p < 0.05) elevations of TNF-α were found in the HEC cultures stimulated with bacterial cells (Pa 8 and Pa 6) compared to the TNF-α levels of the HEC cultures exposed to SBS of Pa 8 and Pa 6. The maximum production of TNF-α was observed at 6 h; after that, TNF-α levels declined dramatically with time. The lowest TNF-α levels were seen 72 h post-exposure to either bacterial cells (Pa 8, Pa 6, and Pa 1) or their SBS. This study demonstrated that P. aeruginosa bacterial cells have a higher ability to stimulate HECs to produce TNF-α than their SBS.
Discussion
This study highlights one of the most important type of cells that contribute to the innate immune response. In addition to being a major barrier that prevents the entry of pathogenic microorganisms, HECs play a pivotal role in the primary immune response, as well as in the inflammatory immune response and phagocytosis.12 Moreover, HECs have a strong ability to produce antibacterial agents (antimicrobial peptides) that prevent pathogenic bacteria from invading the respiratory system.18
This study involved isolating epithelial cells from healthy donors and then culturing them in RPMI medium and exposing them to 3 P. aeruginosa bacterial isolates, as well as stimulating them in vitro with SBS from 3 P. aeruginosa isolates. The study showed for the first time that secretions (SBS) of a bacteria (P. aeruginosa) in growth medium stimulated oral HECs to produce IL-1β and TNF-α. However, the ability of SBS to stimulate the epithelial cells to secrete pro-inflammatory cytokines was limited and less pronounced than the ability of P. aeruginosa cells to stimulate epithelial cells. This supports the notion that bacterial extracts may be used to stimulate the inflammatory immune response and thereby contribute to enhancing the innate immune response against external pathogens nonspecifically.
The ability of P. aeruginosa to stimulate a pro-inflammatory immune response depends on the ability of its surface proteins, as well as flagella and pili, to enhance the immune response of epithelial cells by stimulating them to produce pro-inflammatory cytokines.7 The surface receptors of epithelial cells (i.e., Toll-like receptors (TLRs)) have a role in stimulating these cells to respond to bacterial cell proteins. The TLRs bind to pathogen-associated molecular patterns (PAMPs) and thus are one of the pattern recognition receptors. Toll-like receptor 4 (TLR4) binds to flagellin (the structural protein of flagella), which stimulates the cascades of epithelial cell immune responses that ultimately produce pro-inflammatory cytokines.8 A similar process occurs when epithelial cells are stimulated by other proteins and other materials produced during P. aeruginosa growth in vitro. Molecules that are produced or secreted from bacterial cells or produced when bacterial cells are destroyed, such as nucleic acids and cell membrane lipopolysaccharides, and other enzymes that are produced by bacterial cells during bacterial growth (PAMPs), activate the innate immune system by binding with specific TLRs. This activates different pathways to produce different pro-inflammatory cytokines and ultimately activates the nonspecific immune response against external pathogens.20 Secretions from bacterial cells in growth medium consist of different compounds, and these compounds can stimulate epithelial cells to respond to them. A previous study reported that both intracellular and extracellular sensing play important roles in stimulating the pro-inflammatory immune response via activation of macrophages and other innate immune response arms. This happens through activation of the cellular activation pathway of human cells.21
The results of this study are very interesting, as SBS could be used to stimulate the pro-inflammatory immune response, which may lead to a specific immune response when used with a particular vaccine. The study opens the door to the possibility of using SBS as an adjuvant, which would be better than using the whole bacterial body because the bacterial body can cause severe and prolonged inflammatory responses that have negative effects on lung tissue.22 In contrast, in this study, SBS stimulated the pro-inflammatory immune response in a moderate manner and for a limited period, which is what is required to develop a specific immune response when it combines with a particular antigen. This may help improve the immune response against high-risk pathogens (viruses and bacteria). Our laboratory is conducting several experiments concerning the possibility of using this extract to develop a specific immune response against pathogens. We have also conducted several experiments to determine the ability of SBS to stimulate a nonspecific and pro-inflammatory immune response in an animal model. The positive results we have obtained demonstrate that SBS has the ability to stimulate the mucosal innate immune response in mouse lungs (data not yet published). Several previous studies have highlighted the important role of molecules produced during bacterial growth (i.e., quorum-sensing bacteria produce and release chemical signal molecules called autoinducers). These molecules play a central role in biofilm formation and also play a role in the regulation of virulence bacterial factors.23 In contrast, other studies have shown that nitric oxide produced by P. aeruginosa during bacterial growth negatively affected biofilm structure, which helped it dispose of biofilm mass.24
Conclusions
It can be concluded that the extracellular products released in the growth medium after inoculation with P. aeruginosa successfully promoted pro-inflammatory cytokine production by HECs in vitro. This phenomenon could be implemented by researchers to yield a safe, effective and affordable pro-inflammatory stimulant that could help augment the immune response against external pathogens, both specifically and nonspecifically.